Main research interests: RNAi and Cancer Therapeutics
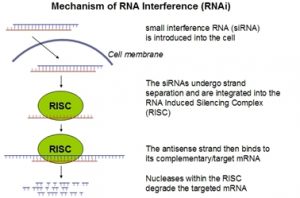
RNA interference (RNAi), a form of post-transcriptional gene silencing induced by introduction of double-stranded RNA (dsRNA), has become a powerful experimental tool for studying gene function. The RNAi phenomenon was first discovered in Caenorhabditis elegans and is characterized by sequence-specific gene silencing elicited by introduction of dsRNA (Fire et al. 1998; Elbashir et al. 2001) complementary to a target mRNA. In the endogenous RNAi pathway, long dsRNA is cleaved by the RNase III type endonuclease, Dicer, to produce 21–23 base pair (bp) short interfering RNAs. The siRNAs are in turn unwound and incorporated into a multiprotein complex known as the RNA-induced silencing complex (RISC), generating a sequence-specific nuclease that guides the cleavage of specific complementary mRNAs. In mammalian cells, direct introduction of siRNAs is used to experimentally initiate RNAi, because introduction of long dsRNA induces a potent antiviral response in addition to RNAi.
Highlight on RNAi research interests: Enabling genome-wide siRNA library screening in cancer cell lines and primary cultures was published in RNA journal (2005) High-throughput RNAi screening in vitro: from cell lines to primary cells. (Ovcharenko et al), followed by genome-wide siRNA and microRNA libraries screening for studying TRAIL-induced apoptosis pathway published in Cancer Research journal (2007) Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. (Ovcharenko et al). Followed by studying microRNA anti-cancer therapeutic applications published in Experimental Hematology journal (2011) miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. (Ovcharenko et al), and siRNA therapeutic applications published in International Journal of Pharmaceutics (2012) Calcium condensed cell penetrating peptide complexes offer highly efficient, low toxicity gene silencing. (Baoum et al).
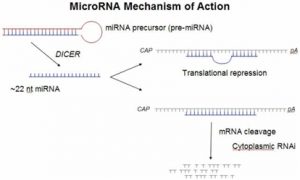
MicroRNAs (miRNAs) are small RNA molecules encoded in the genomes of plants and animals. These newly identified molecules are highly conserved RNAs, up to 22 nucleotides in length, that regulate the expression of genes by binding to the 3′-untranslated regions (3′-UTR) of specific mRNAs. Recent studies of miRNA expression implicate miRNAs in brain development, chronic lymphocytic leukemia, colonic adenocarcinoma, Burkitt’s Lymphoma, and viral infection suggesting possible links between miRNAs and viral disease, neurodevelopment, and cancer. Application of microRNAs as therapeutic targets represent a novel molecular based approach for developing new medicines. While siRNA molecules can target only a single gene for disease treatment, microRNA-based therapeutics will have an advantage of a single microRNA targeting a network of genes with minimal off-target side effects, since miRNAs are naturally expressed in human cells.
RNAi followed by CRISPR. The CRISPR/Cas9 gene editing mechanism is based on a natural defense mechanism prevalent among many bacteria against viruses. CRISPR, which stands for Clustered Regularly Interspersed Short Palindromic Repeats and refers to short, palindromic segments of viral DNA. When a secondary viral infection occurs, a nuclease guided by RNA from the stored CRISPR locates and cleaves the viral DNA. The nuclease, called Cas, has been harnessed for genetic engineering purposes by selecting the sequence that it targets. When double stranded DNA is cut, several repair mechanisms are initiated by the cell. The non-homologous end joining pathway (NHEJ) puts back together the cleaved DNA, which may result in genes being knocked out as a result of being cut off from the main strand. The homology directed repair (HDR) pathway uses nearby strands of DNA that overlap the hanging ends to precisely repair the DNA. HDR can be used to introduce genes into DNA by flagging the desired introduced gene with segments that overlap the cleaved ends of cellular DNA, resulting in a segment known as a repair template. The two above mechanisms can be used to delete desired genes and introduce new ones into cellular DNA.
Novel brain cancer therapeutics described in a recent study published in Cancer Biology and Therapy journal (2019) Two dichloric compounds inhibit in vivo U87 xenograft tumor growth (Ovcharenko et al). This study of DCAH and DCMAH is still ongoing, and active research is being performed to further study mechanism of action, compounds efficacy for various cancer and safety (toxicology). The positive results in a preclinical setting urge further studies that would likewise build up on previous results. Of course – getting a drug on the market is a feat that takes years, if not decades.